If you got samflags and want to know their meaning quickly then you can check their meaning interactively following this link. You type in a flag and will get the answer at Piacrd official site.
You can also look up the meaning of samflags in a pdf file sam_output in pdf. Or it looks like this:
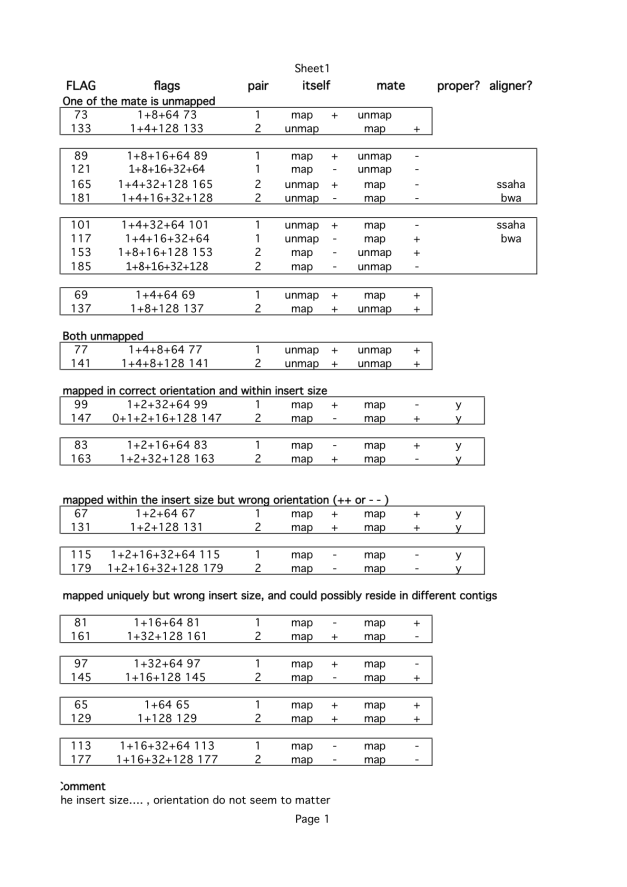 (Provided by WTSI Pathogen Unit). If you do not see the flag you are after, then you can check the picard page or go further below to get actual individual flag components. Or you can just forget about it as an error and check aligners.
(Provided by WTSI Pathogen Unit). If you do not see the flag you are after, then you can check the picard page or go further below to get actual individual flag components. Or you can just forget about it as an error and check aligners.
Please notice positional order of the paired reads “matters” if they are pointing inward or outward.
That is, if you see
A)
1.==> 2.==>
1.<== 2.<==
then the flags are same, since
B)
2.==> 1.==>
2.<== 1.<==
are exactly same to A).
But,
1.==> 2.<==
is not same as
2.<== 1.==>
or vice versa.
This can indicate tandem or episomal duplications or errors. If reads are overlapping, then you see this quite often. (Even experts may not know this. It took me more than 30 minutes convince people.) If you are in doubt, please check all read flags carefully. You may not see them since they can be very rare, which does not mean they do not exist.
It is not easy to identify structural variation (SV) solely based on insert sizes. People unaware of this difficulty think they can just find SVs from their insert sizes: Sorry, but it is not that simple. I was able to find only extremely tandemly repeated gene arrays with this method. Especially it is almost impossible to do this for a draft reference containing many errors.
Here is the flags you should expect after mapping. They are insert size. Note: the orientation of some reads is not defined, and I do not know why. To analyse the flags properly, you may have to order bam files by their name, since you need to know the mapping status of mates. For instance, you need to know both sam flags and positional arrangement of reads. Insert size can help to throw away badly mapped reads but identifying valid SVs is really tough. Or the parasites I am working on may not have very many SVs.
Please check the 1000G paper to find SV detection. The paper A map of human genome variation from population-scale sequencing is available from the Nature web site and is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence to ensure wide distribution.
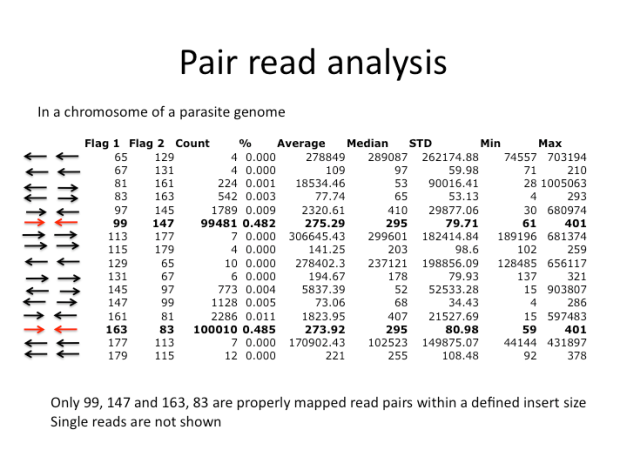 There are many errors in this figure. The direction is not defined in 65 and 129. I need to fix this.
There are many errors in this figure. The direction is not defined in 65 and 129. I need to fix this.
Structural Variant based on Apparent Insert Size:
Small insertion => Decrease
Small deletion => Increase
Large deletion => Increase
Intrachromosomal translocation => Increase
Interchromosomal translocation => Change mate chromosome
Large insertion => Increase or lack of mapped mate
From: Finding Structural Variants in Short Read, Paired-end Sequence Data with R and Bioconductor by Sean Davis
General Information
http://samtools.sourceforge.net/
- SAM Format Specification
- SF Project Page
- SF Download Page
- Mailing Lists
- SVN Browse
- Related Software
- FAQ
Other Lang-bindings
- BamTools (C++)
- Picard (Java)
- Bio-SamTools (Perl)
- Pysam (Python)
- Samtools-Ruby (Ruby)
- cl-sam (Common Lisp)
Or more generally they are
0x0001 1 the read is paired in sequencing, no matter whether it is mapped in a pair
0x0002 2 the read is mapped in a proper pair (depends on the protocol, normally inferred during alignment)
0x0004 4 the query sequence itself is unmapped
0x0008 8 the mate is unmapped
0x0010 16 strand of the query (0 for forward; 1 for reverse strand)
0x0020 32 strand of the mate
0x0040 64 the read is the first read in a pair
0x0080 128 the read is the second read in a pair
0x0100 256 the alignment is not primary (a read having split hits may have multiple primary alignment records)
========== PrintSamFlag.pl : Usage perl PrintSamFlag.pl input.bam > output
# This program will print out a flag status like this
64706 65 ‘1”64’
61396 69 ‘1”4”64’
66631 73 ‘1”8”64’
130783 77 ‘1”4”8”64’
151001 81 ‘1”16”64’
4225910 83 ‘1”2”16”64’
63678 89 ‘1”8”16”64’
151042 97 ‘1”32”64’
4188825 99 ‘1”2”32”64’
61159 101 ‘1”4”32”64’
63276 113 ‘1”16”32”64’
64707 129 ‘1”128’
66631 133 ‘1”4”128’
61396 137 ‘1”8”128’
130783 141 ‘1”4”8”128’
151042 145 ‘1”16”128’
4200609 147 ‘1”2”16”128’
61159 153 ‘1”8”16”128’
151001 161 ‘1”32”128’
4175327 163 ‘1”2”32”128’
63678 165 ‘1”4”32”128’
63276 177 ‘1”16”32”128’
=======Please copy and paste the following lines and check they are pasted correctly.===========
#!/usr/bin/perl
use strict;
my ($in)=@ARGV;
if ($in=~"\.bam"){
open F1," samtools view $in |" or die;
}
if ($in=~"\.sam"){
open F1,$in or die;
}
my (%flag,%INS,$n1,$n2,@x,$InsS,$P);
while (my $s=<F1>)
{
@x=split '\s+',$s;
++$flag{$x[1]};
++$INS{$x[1]};
#paired
if (SAMflag($x[1])=~"\'2\'"){
#print SAMflag($x[1]) ,"\n";
$InsS+=$x[8];
++$P;
}
}
close F1;
for my $k (sort {$a<=>$b}keys %flag){
print "$flag{$k} $k ",SAMflag($k),"\n";
}
my %TABLE= qw( 0 '0'
1 '1'
2 '2'
3 '1''2'
4 '4'
5 '1''4'
6 '2''4'
7 '1''2''4'
8 '8'
9 '1''8'
10 '2''8'
11 '1''2''8'
12 '4''8'
13 '1''4''8'
14 '2''4''8'
15 '1''2''4''8'
16 '16'
17 '1''16'
18 '2''16'
19 '1''2''16'
20 '4''16'
21 '1''4''16'
22 '2''4''16'
23 '1''2''4''16'
24 '8''16'
25 '1''8''16'
26 '2''8''16'
27 '1''2''8''16'
28 '4''8''16'
29 '1''4''8''16'
30 '2''4''8''16'
31 '1''2''4''8''16'
32 '32'
33 '1''32'
34 '2''32'
35 '1''2''32'
36 '4''32'
37 '1''4''32'
38 '2''4''32'
39 '1''2''4''32'
40 '8''32'
41 '1''8''32'
42 '2''8''32'
43 '1''2''8''32'
44 '4''8''32'
45 '1''4''8''32'
46 '2''4''8''32'
47 '1''2''4''8''32'
48 '16''32'
49 '1''16''32'
50 '2''16''32'
51 '1''2''16''32'
52 '4''16''32'
53 '1''4''16''32'
54 '2''4''16''32'
55 '1''2''4''16''32'
56 '8''16''32'
57 '1''8''16''32'
58 '2''8''16''32'
59 '1''2''8''16''32'
60 '4''8''16''32'
61 '1''4''8''16''32'
62 '2''4''8''16''32'
63 '1''2''4''8''16''32'
64 '64'
65 '1''64'
66 '2''64'
67 '1''2''64'
68 '4''64'
69 '1''4''64'
70 '2''4''64'
71 '1''2''4''64'
72 '8''64'
73 '1''8''64'
74 '2''8''64'
75 '1''2''8''64'
76 '4''8''64'
77 '1''4''8''64'
78 '2''4''8''64'
79 '1''2''4''8''64'
80 '16''64'
81 '1''16''64'
82 '2''16''64'
83 '1''2''16''64'
84 '4''16''64'
85 '1''4''16''64'
86 '2''4''16''64'
87 '1''2''4''16''64'
88 '8''16''64'
89 '1''8''16''64'
90 '2''8''16''64'
91 '1''2''8''16''64'
92 '4''8''16''64'
93 '1''4''8''16''64'
94 '2''4''8''16''64'
95 '1''2''4''8''16''64'
96 '32''64'
97 '1''32''64'
98 '2''32''64'
99 '1''2''32''64'
100 '4''32''64'
101 '1''4''32''64'
102 '2''4''32''64'
103 '1''2''4''32''64'
104 '8''32''64'
105 '1''8''32''64'
106 '2''8''32''64'
107 '1''2''8''32''64'
108 '4''8''32''64'
109 '1''4''8''32''64'
110 '2''4''8''32''64'
111 '1''2''4''8''32''64'
112 '16''32''64'
113 '1''16''32''64'
114 '2''16''32''64'
115 '1''2''16''32''64'
116 '4''16''32''64'
117 '1''4''16''32''64'
118 '2''4''16''32''64'
119 '1''2''4''16''32''64'
120 '8''16''32''64'
121 '1''8''16''32''64'
122 '2''8''16''32''64'
123 '1''2''8''16''32''64'
124 '4''8''16''32''64'
125 '1''4''8''16''32''64'
126 '2''4''8''16''32''64'
127 '1''2''4''8''16''32''64'
128 '128'
129 '1''128'
130 '2''128'
131 '1''2''128'
132 '4''128'
133 '1''4''128'
134 '2''4''128'
135 '1''2''4''128'
136 '8''128'
137 '1''8''128'
138 '2''8''128'
139 '1''2''8''128'
140 '4''8''128'
141 '1''4''8''128'
142 '2''4''8''128'
143 '1''2''4''8''128'
144 '16''128'
145 '1''16''128'
146 '2''16''128'
147 '1''2''16''128'
148 '4''16''128'
149 '1''4''16''128'
150 '2''4''16''128'
151 '1''2''4''16''128'
152 '8''16''128'
153 '1''8''16''128'
154 '2''8''16''128'
155 '1''2''8''16''128'
156 '4''8''16''128'
157 '1''4''8''16''128'
158 '2''4''8''16''128'
159 '1''2''4''8''16''128'
160 '32''128'
161 '1''32''128'
162 '2''32''128'
163 '1''2''32''128'
164 '4''32''128'
165 '1''4''32''128'
166 '2''4''32''128'
167 '1''2''4''32''128'
168 '8''32''128'
169 '1''8''32''128'
170 '2''8''32''128'
171 '1''2''8''32''128'
172 '4''8''32''128'
173 '1''4''8''32''128'
174 '2''4''8''32''128'
175 '1''2''4''8''32''128'
176 '16''32''128'
177 '1''16''32''128'
178 '2''16''32''128'
179 '1''2''16''32''128'
180 '4''16''32''128'
181 '1''4''16''32''128'
182 '2''4''16''32''128'
183 '1''2''4''16''32''128'
184 '8''16''32''128'
185 '1''8''16''32''128'
186 '2''8''16''32''128'
187 '1''2''8''16''32''128'
188 '4''8''16''32''128'
189 '1''4''8''16''32''128'
190 '2''4''8''16''32''128'
191 '1''2''4''8''16''32''128'
192 '64''128'
193 '1''64''128'
194 '2''64''128'
195 '1''2''64''128'
196 '4''64''128'
197 '1''4''64''128'
198 '2''4''64''128'
199 '1''2''4''64''128'
200 '8''64''128'
201 '1''8''64''128'
202 '2''8''64''128'
203 '1''2''8''64''128'
204 '4''8''64''128'
205 '1''4''8''64''128'
206 '2''4''8''64''128'
207 '1''2''4''8''64''128'
208 '16''64''128'
209 '1''16''64''128'
210 '2''16''64''128'
211 '1''2''16''64''128'
212 '4''16''64''128'
213 '1''4''16''64''128'
214 '2''4''16''64''128'
215 '1''2''4''16''64''128'
216 '8''16''64''128'
217 '1''8''16''64''128'
218 '2''8''16''64''128'
219 '1''2''8''16''64''128'
220 '4''8''16''64''128'
221 '1''4''8''16''64''128'
222 '2''4''8''16''64''128'
223 '1''2''4''8''16''64''128'
224 '32''64''128'
225 '1''32''64''128'
226 '2''32''64''128'
227 '1''2''32''64''128'
228 '4''32''64''128'
229 '1''4''32''64''128'
230 '2''4''32''64''128'
231 '1''2''4''32''64''128'
232 '8''32''64''128'
233 '1''8''32''64''128'
234 '2''8''32''64''128'
235 '1''2''8''32''64''128'
236 '4''8''32''64''128'
237 '1''4''8''32''64''128'
238 '2''4''8''32''64''128'
239 '1''2''4''8''32''64''128'
240 '16''32''64''128'
241 '1''16''32''64''128'
242 '2''16''32''64''128'
243 '1''2''16''32''64''128'
244 '4''16''32''64''128'
245 '1''4''16''32''64''128'
246 '2''4''16''32''64''128'
247 '1''2''4''16''32''64''128'
248 '8''16''32''64''128'
249 '1''8''16''32''64''128'
250 '2''8''16''32''64''128'
251 '1''2''8''16''32''64''128'
252 '4''8''16''32''64''128'
253 '1''4''8''16''32''64''128'
254 '2''4''8''16''32''64''128'
255 '1''2''4''8''16''32''64''128');
sub SAMflag{
my $i=shift;
return $TABLE{$i};
}
#======= END ====================
SAMtool bitwise flag: A table of flags with combination of integers in perl format. There is a better way to do this but it is useful, for some.:)
use strict; my %TABLE= qw(0 0 1 1 2 2 3 1+2 4 0+4 5 1+4 6 0+2+4 7 1+2+4 8 0+8 9 1+8 10 0+2+8 11 1+2+8 12 0+4+8 13 1+4+8 14 0+2+4+8 15 1+2+4+8 16 0+16 17 1+16 18 0+2+16 19 1+2+16 20 0+4+16 21 1+4+16 22 0+2+4+16 23 1+2+4+16 24 0+8+16 25 1+8+16 26 0+2+8+16 27 1+2+8+16 28 0+4+8+16 29 1+4+8+16 30 0+2+4+8+16 31 1+2+4+8+16 32 0+32 33 1+32 34 0+2+32 35 1+2+32 36 0+4+32 37 1+4+32 38 0+2+4+32 39 1+2+4+32 40 0+8+32 41 1+8+32 42 0+2+8+32 43 1+2+8+32 44 0+4+8+32 45 1+4+8+32 46 0+2+4+8+32 47 1+2+4+8+32 48 0+16+32 49 1+16+32 50 0+2+16+32 51 1+2+16+32 52 0+4+16+32 53 1+4+16+32 54 0+2+4+16+32 55 1+2+4+16+32 56 0+8+16+32 57 1+8+16+32 58 0+2+8+16+32 59 1+2+8+16+32 60 0+4+8+16+32 61 1+4+8+16+32 62 0+2+4+8+16+32 63 1+2+4+8+16+32 64 0+64 65 1+64 66 0+2+64 67 1+2+64 68 0+4+64 69 1+4+64 70 0+2+4+64 71 1+2+4+64 72 0+8+64 73 1+8+64 74 0+2+8+64 75 1+2+8+64 76 0+4+8+64 77 1+4+8+64 78 0+2+4+8+64 79 1+2+4+8+64 80 0+16+64 81 1+16+64 82 0+2+16+64 83 1+2+16+64 84 0+4+16+64 85 1+4+16+64 86 0+2+4+16+64 87 1+2+4+16+64 88 0+8+16+64 89 1+8+16+64 90 0+2+8+16+64 91 1+2+8+16+64 92 0+4+8+16+64 93 1+4+8+16+64 94 0+2+4+8+16+64 95 1+2+4+8+16+64 96 0+32+64 97 1+32+64 98 0+2+32+64 99 1+2+32+64 100 0+4+32+64 101 1+4+32+64 102 0+2+4+32+64 103 1+2+4+32+64 104 0+8+32+64 105 1+8+32+64 106 0+2+8+32+64 107 1+2+8+32+64 108 0+4+8+32+64 109 1+4+8+32+64 110 0+2+4+8+32+64 111 1+2+4+8+32+64 112 0+16+32+64 113 1+16+32+64 114 0+2+16+32+64 115 1+2+16+32+64 116 0+4+16+32+64 117 1+4+16+32+64 118 0+2+4+16+32+64 119 1+2+4+16+32+64 120 0+8+16+32+64 121 1+8+16+32+64 122 0+2+8+16+32+64 123 1+2+8+16+32+64 124 0+4+8+16+32+64 125 1+4+8+16+32+64 126 0+2+4+8+16+32+64 127 1+2+4+8+16+32+64 128 0+128 129 1+128 130 0+2+128 131 1+2+128 132 0+4+128 133 1+4+128 134 0+2+4+128 135 1+2+4+128 136 0+8+128 137 1+8+128 138 0+2+8+128 139 1+2+8+128 140 0+4+8+128 141 1+4+8+128 142 0+2+4+8+128 143 1+2+4+8+128 144 0+16+128 145 1+16+128 146 0+2+16+128 147 1+2+16+128 148 0+4+16+128 149 1+4+16+128 150 0+2+4+16+128 151 1+2+4+16+128 152 0+8+16+128 153 1+8+16+128 154 0+2+8+16+128 155 1+2+8+16+128 156 0+4+8+16+128 157 1+4+8+16+128 158 0+2+4+8+16+128 159 1+2+4+8+16+128 160 0+32+128 161 1+32+128 162 0+2+32+128 163 1+2+32+128 164 0+4+32+128 165 1+4+32+128 166 0+2+4+32+128 167 1+2+4+32+128 168 0+8+32+128 169 1+8+32+128 170 0+2+8+32+128 171 1+2+8+32+128 172 0+4+8+32+128 173 1+4+8+32+128 174 0+2+4+8+32+128 175 1+2+4+8+32+128 176 0+16+32+128 177 1+16+32+128 178 0+2+16+32+128 179 1+2+16+32+128 180 0+4+16+32+128 181 1+4+16+32+128 182 0+2+4+16+32+128 183 1+2+4+16+32+128 184 0+8+16+32+128 185 1+8+16+32+128 186 0+2+8+16+32+128 187 1+2+8+16+32+128 188 0+4+8+16+32+128 189 1+4+8+16+32+128 190 0+2+4+8+16+32+128 191 1+2+4+8+16+32+128 192 0+64+128 193 1+64+128 194 0+2+64+128 195 1+2+64+128 196 0+4+64+128 197 1+4+64+128 198 0+2+4+64+128 199 1+2+4+64+128 200 0+8+64+128 201 1+8+64+128 202 0+2+8+64+128 203 1+2+8+64+128 204 0+4+8+64+128 205 1+4+8+64+128 206 0+2+4+8+64+128 207 1+2+4+8+64+128 208 0+16+64+128 209 1+16+64+128 210 0+2+16+64+128 211 1+2+16+64+128 212 0+4+16+64+128 213 1+4+16+64+128 214 0+2+4+16+64+128 215 1+2+4+16+64+128 216 0+8+16+64+128 217 1+8+16+64+128 218 0+2+8+16+64+128 219 1+2+8+16+64+128 220 0+4+8+16+64+128 221 1+4+8+16+64+128 222 0+2+4+8+16+64+128 223 1+2+4+8+16+64+128 224 0+32+64+128 225 1+32+64+128 226 0+2+32+64+128 227 1+2+32+64+128 228 0+4+32+64+128 229 1+4+32+64+128 230 0+2+4+32+64+128 231 1+2+4+32+64+128 232 0+8+32+64+128 233 1+8+32+64+128 234 0+2+8+32+64+128 235 1+2+8+32+64+128 236 0+4+8+32+64+128 237 1+4+8+32+64+128 238 0+2+4+8+32+64+128 239 1+2+4+8+32+64+128 240 0+16+32+64+128 241 1+16+32+64+128 242 0+2+16+32+64+128 243 1+2+16+32+64+128 244 0+4+16+32+64+128 245 1+4+16+32+64+128 246 0+2+4+16+32+64+128 247 1+2+4+16+32+64+128 248 0+8+16+32+64+128 249 1+8+16+32+64+128 250 0+2+8+16+32+64+128 251 1+2+8+16+32+64+128 252 0+4+8+16+32+64+128 253 1+4+8+16+32+64+128 254 0+2+4+8+16+32+64+128 255 1+2+4+8+16+32+64+128);

Flag for properly paired reads.
samtools view -f 2
will give you properly mapped pair.
Usage: samtools view [options] | [region1 […]]
Options: -b output BAM
-h print header for the SAM output
-H print header only (no alignments)
-S input is SAM
-u uncompressed BAM output (force -b)
-x output FLAG in HEX (samtools-C specific)
-X output FLAG in string (samtools-C specific)
-t FILE list of reference names and lengths (force -S) [null]
-T FILE reference sequence file (force -S) [null]
-o FILE output file name [stdout]
-f INT required flag, 0 for unset [0]
-F INT filtering flag, 0 for unset [0]
-q INT minimum mapping quality [0]
-l STR only output reads in library STR [null]
-r STR only output reads in read group STR [null]
-? longer help
Thanks, it help me a lot 🙂
Thanks for the table. I think that will be useful.
Hello, need help for samtools or other methods.
You know that the Sequence Alignment/Map (SAM) format is a generic alignment format for storing read alignments against reference sequences. If a patient’s BAM file is given, how can you extract the sequence from the storing read alignments(not from the reference) within a region? For example, chr22: 1000000-1234000.
Somebody in an online forum totally misunderstood my question and I haven’t get a solution. So I ask for help whereever it is possible. The guy used “samtools faidx ref.fasta”, but this result from the reference instead of the patient’s read.
Thanks for any hint or a piece of code.
Thanks for this post. It was definitely helpful. I was wondering if you could elaborate a little more on the:
“the alignment is not primary (a read having split hits may have multiple primary alignment records)”
Does this mean that if a read has multiple alignments, then one of the alignments is considered the primary alignment? How does this primary alignment get assigned? Is it based on the best of all the alignments?
Thanks,
ardmore: If a patient’s BAM file is given, how can you extract the sequence from the storing read alignments(not from the reference) within a region? For example, chr22: 1000000-1234000.
Sorry I did not check this for a while. It is indeed tricky: you need reverse complement reverse reads: 1 st mates.
Here is bam2 fastq for a given region.
perl Bam2fastqR.pl my.bam chr_name start end
perl Bam2fastqR.pl my.bam chr01 1000 3000
makes
my.2.fastq
###Bam2fastqR.pl (For reverse reads )=======================================
use strict;
use Cwd;
my ($in,$chr,$sta,$end)=@ARGV;
open F1,”samtools view -f 64 $in ‘$chr\:$sta\-$end’ |” or die;
my (@x,@z,$pr1,$pr2,$rb,$rs);
my (@y)=split ‘\.’,$in;
open R1, “>$y[0].1.fastq”;
while (my $s=){
(@x)=split ‘\s+’,$s;
$rs=revdnacomp($x[9]);
$rb=reverse($x[10]);# you need to reverse complement
print R1 “\@$x[0]/1\n$rs\n+\n$rb\n”;
}
sub revdnacomp {
my $dna = shift;
my $revcomp = reverse($dna);
$revcomp =~ tr/ACGTacgt/TGCAtgca/;
return $revcomp;
}
#### END =======================================
###Bam2fastqF.pl (For forward reads )
use strict;
use Cwd;
my ($in,$chr,$sta,$end)=@ARGV;
open F1,”samtools view -f 128 $in ‘$chr\:$sta\-$end’ |” or die;
my (@x,@z,$pr1,$pr2);
my (@y)=split ‘\.’,$in;
open R1, “>$y[0].2.fastq”;
while (my $s=){
(@x)=split ‘\s+’,$s;
print R1 “\@$x[0]/2\n$x[9]\n+\n$x[10]\n”;
}
================================================
You need to run for both scripts. If they do not work, please let me know.
“the alignment is not primary (a read having split hits may have multiple primary alignment records)”
Does this mean that if a read has multiple alignments, then one of the alignments is considered the primary alignment? How does this primary alignment get assigned? Is it based on the best of all the alignments?
============
I just copied it from the samtools manual. And the updated one tells us that
“Bit 0x100 marks the alignment not to be used in certain analyses when the tools in use are
aware of this bit.” So basically we should not use this one, unless an aligner specifically say we should use this flag. I talked with Heng before and he said this flag was to eliminate bad reads. (or something like that.)
please see http://seqanswers.com/forums/showthread.php?t=15459
I borrowed the code from http://edwards.sdsu.edu/labsite/index.php/robert/289-how-to-convert-fastq-to-fasta
But I got something in my fasta file like this, is it right?
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
f[“c`_]d]aWi]c]dbgfi[aZ_d_n_nr[`jlvnmdimUbmb]mfUkkjg^faZ[ih
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~{~~~q~wq[\SVT\]]X`_a_c^XZZYT
`zdsobkpRNRqx}l~`yoyvwo\c~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
zxtuu~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~zsoml\R
I think you are getting something like this. The first part is base and the second part is its quality.
@Ld01_v01s1
CTAACCCTAACCCTAACCCTAACCCTAACCAGYACACCAGTACACCGTCACGCCCCCGTC
CTGTTGGAGAGGGTGTCGCKGTGCAAGGAATCAGTCGAGAGAAAAACCCTAACCCGTACC
GGTACCGAGTTATCGTTTTTATTAGGTTACTAACCTCTGGCACGCTTGTCGCTGCTCTTC
AGAACGAAACGCACAATGCTCTTCGATAAACGTGCTGAAATAGAAAAAAAAAACGAAAGA
TCTCGGCTACGTTTGCTGCCGTTGGCCTCACCTCGTCATGGCGCGTCGGTCCACCAAAGA
+
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~d~~~~~~~~~~~~~~~~~~~~~~~~~~~
~~~~~~~~~~~~~~~~~~~O~~~~~~~~~~~~~~~:~~~~~~~~~~~~~~~~~~~~~~~~
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~~
Hi, just a short Q, how would I select alignments with flag 0 OR 16 using samtools?
A:
samtools view -f 16
0 + 16 => 16
Usage: samtools view [options] | [region1 […]]
Options: -b output BAM
-h print header for the SAM output
-H print header only (no alignments)
-S input is SAM
-u uncompressed BAM output (force -b)
-1 fast compression (force -b)
-x output FLAG in HEX (samtools-C specific)
-X output FLAG in string (samtools-C specific)
-c print only the count of matching records
-L FILE output alignments overlapping the input BED FILE [null]
-t FILE list of reference names and lengths (force -S) [null]
-T FILE reference sequence file (force -S) [null]
-o FILE output file name [stdout]
-R FILE list of read groups to be outputted [null]
-f INT required flag, 0 for unset [0]
-F INT filtering flag, 0 for unset [0]
-q INT minimum mapping quality [0]
-l STR only output reads in library STR [null]
-r STR only output reads in read group STR [null]
-s FLOAT fraction of templates to subsample; integer part as seed [-1]
-? longer help
how would I select alignments with flag 0 OR 16 using samtools?
samtools view -f 16
samtools view -f 0
samtools view -f 17
You needs pick 3 cases, I guess …
Hi! Great post! 🙂
Do you have any idea how to collect the uniquely mapped reads whose 5′ ends falls within a target exon? I’ve filtered for 99, 147, 86 and 163 flags but saddly I don’t have any idea how to get it…
Can someone propose an approach? Thank you in advance!
>Do you have any idea how to collect the uniquely mapped reads whose 5′ ends falls within a target exon? I’ve filtered for 99, 147, 86 and 163 flags but saddly I don’t have any idea how to get it…
It seems manipulating flags is not enough to get the info … Let me see.
I’ve been unsuccessfully struggling with that… because my rookie status I guess.
Maybe filtering from a sam file? If you discard the reads according to the fourth column… should work!
Oh please forget the las comment… I’m a dumbass!